A Comprehensive Toolkit for NLRP3 Inflammasome Drug Discovery
Abstract
The NLR family pyrin domain containing 3 (NLRP3) inflammasome, a key regulator of the innate immune system, mediates caspase-1 activation and the secretion of proinflammatory cytokines such as IL-1β and IL-18 in response to various stimuli. Aberrant NLRP3 function has been implicated in multiple inflammatory diseases, suggesting it as a promising therapeutic target. Conventional methods for measuring NLRP3 activity including immunoassays, fluorescence microscopy, and pull-down assays are handy, but are not effective for establishing a relationship between NLRP3 engagement and pathway inhibition in a cellular environment. In this article, we highlight work from Teske et al., which discusses key findings on inflammasome inhibitors using three scalable cell-based methods providing mechanistic and quantitative information about NLRP3 activation: NLRP3 NanoBRET® Target Engagement (TE) assay, Lumit® IL-1β Immunoassay, and Caspase-Glo® 1 Inflammasome Assay. Together, these assays provide a complementary platform of unique tools that link NLRP3 binding to inhibition of functional inflammasome responses critical for drug discovery workflows.
Introduction
Inflammasomes are cytoplasmic multimeric protein complexes that are vital for proper function of the innate immune system. In response to exogenous pathogens or endogenous danger signals, inflammasomes initiate an inflammation response to protect the host. While there are multiple types of inflammasomes, the NLR family pyrin domain containing 3 (NLRP3) inflammasome is the most well-characterized. In addition to responding to a wide range of stimuli, the NLRP3 inflammasome is involved in both innate and adaptive immunity. Dysregulation of NLRP3 inflammasome activation has been associated with the onset and progression of several diseases including autoimmune disorders and Alzheimer's disease. Thus, research assessing the NLRP3 inflammasome as a therapeutic target is crucial for advancing potential therapeutics in the pipeline.
Recently, a team of Promega research scientists led by Martha O’Brien evaluated the efficacy of seven inhibitors targeting the NLRP3 inflammasome in a Cell Chemical Biology publication using the NLRP3 NanoBRET® Target Engagement (TE) assay, Lumit® IL-1β Immunoassay, and Caspase-Glo® 1 Inflammasome Assay. The team first established that all three assays yielded consistent results for inflammasome activity by using MCC950, a well-characterized and potent inhibitor of the NLRP3 inflammasome. Then they tested the effectiveness of six additional reported NLRP3 inflammasome inhibitors including Oridonin, NBC19, NBC6, CY09, OXSI-2, and OLT1177. Using these three assays, they were able to rank the potency of five published NLRP3 inhibitors and demonstrated that two inhibitors failed to yield inhibitory effects on the NLRP3 inflammasome.
Conventional Methods to Detect NLRP3 Inflammasome Activation
There are many common methods used to quantify inflammasome activity:
Microscopy
Traditional immunofluorescence staining techniques label ASC specks with a specific anti-ASC antibody to measure inflammasome activation. Alternatively, some cell lines can stably express a construct containing ASC-mCerulean, ASC-mCherry or ASC fused to other fluorescent tags, enabling the formation of ASC specks to be observed in real time without staining. While this method is useful for visualizing ASC speck protein expression and localization, staining protocols are lengthy, require multiple wash steps, and do not translate well to high-throughput applications.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs are also used to quantify inflammasome activation, which usually involves measuring the mature forms of IL-1β and IL-18 pro-inflammatory cytokines. Some ELISA kits detect NLRP3 and high mobility group box 1 (HMGB1), which is an effector molecule that is induced by NLRP3 inflammasome activation. ELISA kits measuring NLRP3, however, only detect the protein’s presence and does not report on its activation. Additionally, these assays are long, requiring many steps and introducing more opportunity for error. ELISAs also have a limited linear range and samples often require sample dilution to be detected in the proper range.
Western blots and pull-down assays
Pull-down assays enable inflammasome activation to be monitored through specific protein interactions with the NLRP3 inflammasome and can allow researchers to use recombinant NLRP3 protein to discover the protein’s binding properties. After the pull-down assay, further downstream analysis such as a Western blot is usually warranted to check if the protein of interest has been pulled down together with the target protein. Western blots are also frequently employed to evaluate inflammasome activation through the processing of the pro-inflammatory cytokines including IL-1β and IL-18 as well as Gasdermin D and other various caspases. Both approaches are cumbersome and demand a specialized skillset. Furthermore, classic western blots are nonquantitative and require specific primary antibodies against the protein of interest, which can severely limit detection possibilities.
Assays for Monitoring Inflammasome Activation
Promega offers scalable assays with simple no-wash protocols that provide an easier way to investigate NLRP3 inflammasome activation. With the NLRP3 NanoBRET® TE assay, target engagement of NLRP3 inhibitors can be observed using a fluorescent tracer based on the inhibitor molecule (Figure 1). This assay uses bioluminescent resonance energy transfer (BRET), an energy transfer technique, to measure binding between a target protein and small molecules in live cells. The target engagement assay can be complemented with the Lumit® IL-1β Immunoassay, Lumit® Active IL-18 Immunoassay, and Caspase-Glo® 1 Inflammasome Assay measuring secreted IL-1β and active IL-18 as well as caspase-1 activity, respectively, to link biophysical target engagement with functional outcomes in cells (Figure 2). The Lumit® Immunoassays and Caspase-Glo® 1 Inflammasome Assay all have add-and-read methods, yielding a luminescent readout in 70 minutes or less that is more sensitive than colorimetric or fluorometric measures.
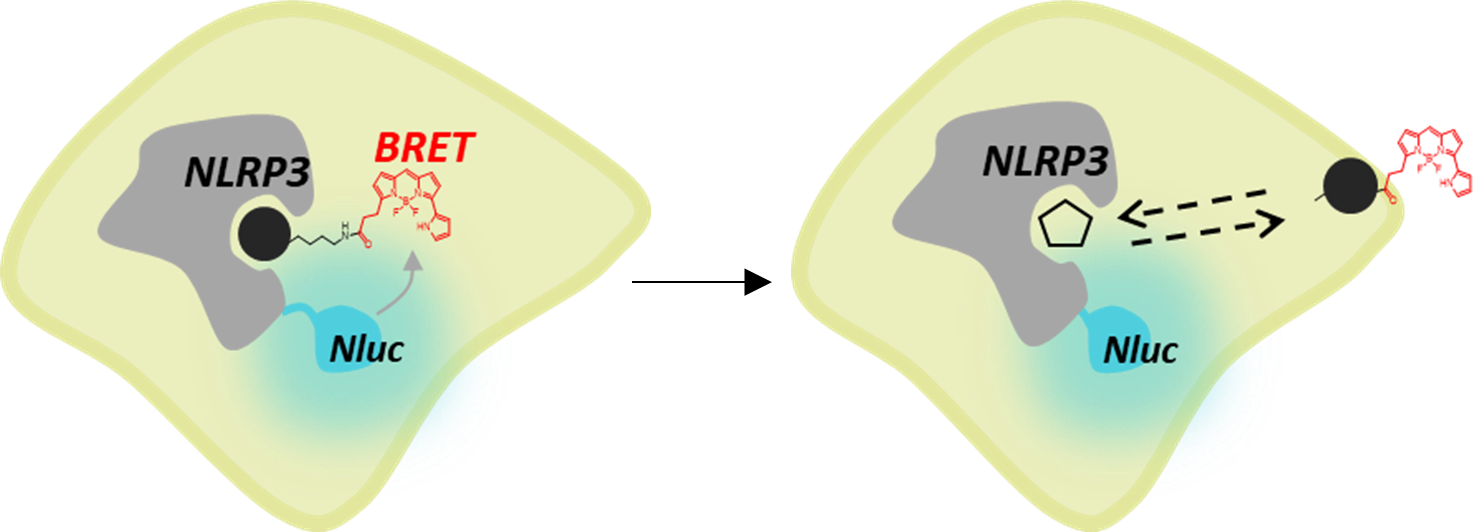
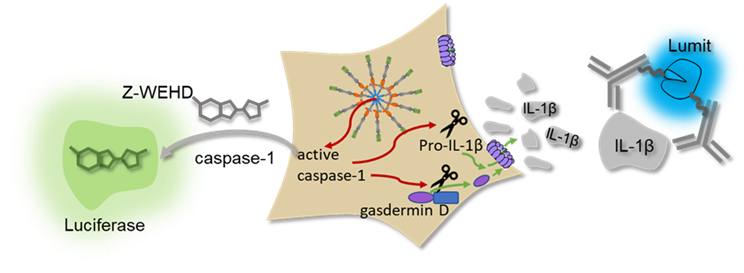
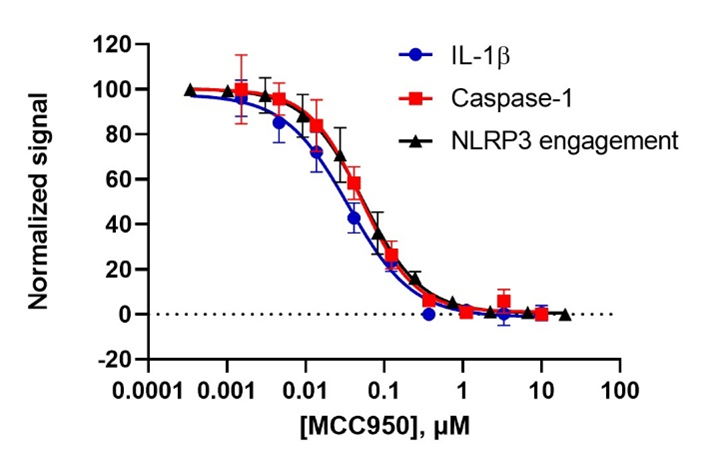
Comparing Rank Potency of Antagonists to the NLRP3 Inflammasome
These three assays were used with other NLRP3 inhibitors including Oridonin, NBC19, NBC6, and CY-09, allowing the rank order potency between these to be established. The NLRP3 target engagement assay was carried out in HEK293 cells while measures of caspase-1 activity and IL-1β secretion were made using lipopolysaccharide (LPS)-primed peripheral blood mononuclear cells (PBMCs) treated with nigericin in a split sample analysis, enabling measurements to be collected from the same cell wells (Figure 4). In all three assays, MCC950 was the most potent of the five compounds. With little difference in their IC50s, the next most potent compounds for the functional assays are Oridonin, NBC19, and NBC6. The least potent compound in all three assays was CY-09.
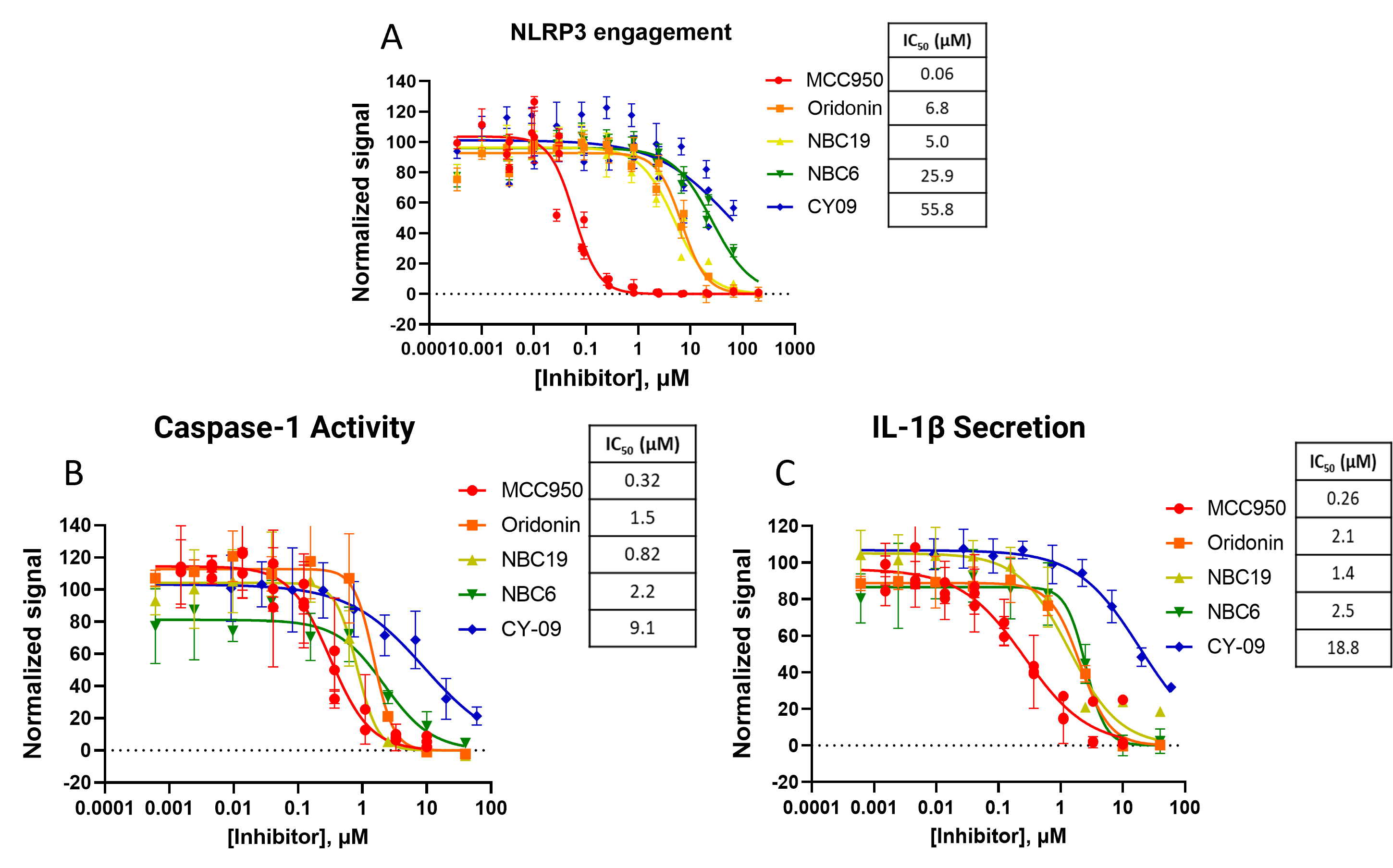
Validation of NLRP3 Inhibition Using Cell-Based Assays
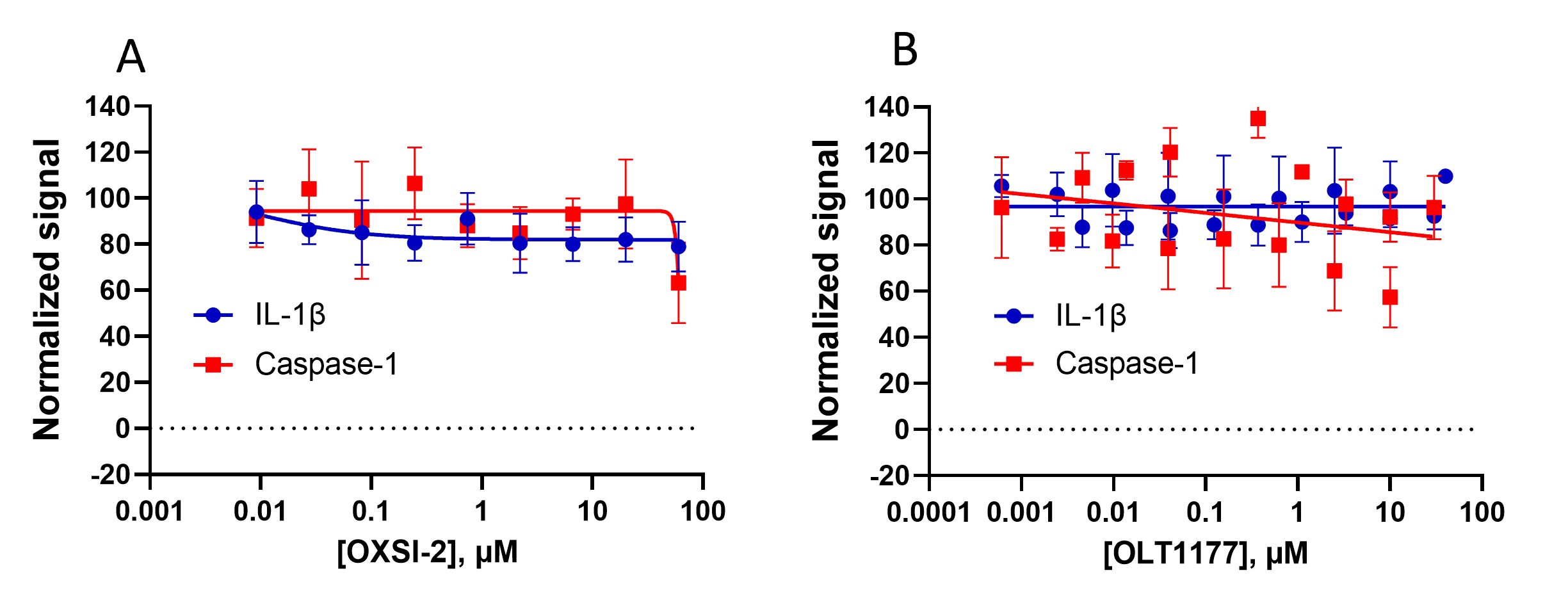
Among the inhibitors Teske et al. evaluated in their recent publication was OXSI-2 and OLT1177, two previously reported NLRP3 inhibitors. Unlike the inhibitors tested in Figure 4, OXSI-2 and OLT1177 did not demonstrate inhibition in any of the functional inflammasome assays (Figure 5). OXSI-2 did not inhibit caspase-1 activity or IL-1β release in inflammasome stimulated PBMCs as indicated by the absence of an inhibition curve (Figure 5A). Similarly, OLT1177 did not show inhibition of functional measures in PBMCs (Figure 5B). The lack of inhibition in these functional assays independent of quenching effects indicates that both OXSI-2 and OLT1177 are not direct NLRP3 inhibitors at concentrations previously used in cell models.
Summary
The NLRP3 NanoBRET® TE Assay, Lumit® IL-1β Immunoassay and Caspase-Glo® 1 Inflammasome Assay provide a comprehensive panel of cell-based assays for researchers who need to query the drug mode of action for the NLRP3 inflammasome. These assays combined are useful for quantifying inflammasome inhibition, measuring specific NLRP3 engagement, and characterizing small molecule NLRP3 inhibitors. All three methods have simple, no-wash protocols with minimal cellular manipulation, enabling compounds to be titrated at broader ranges to achieve a complete dose response.
Additional resources for characterizing inflammasome activation can be found here.
Reference
Teske, K.A. et al. (2024) Interrogating direct NLRP3 engagement and functional inflammasome inhibition using cellular assays. Cell Chem. Biol. 31(2), 349-360.e6.
Find the full publication discussed in this article available through open access at https://www.sciencedirect.com/science/article/pii/S2451945623003355.
Learn more about the development of the Caspase-Glo® 1 Inflammasome Assay.
Related Resources

Webinar: Lumit™ Immunoassays
This webinar covers how Lumit™ Immunoassays are a faster method for analyte detection.
Article: Lumit™ Cytokine Assays
Learn how to Interpolate Data with the GloMax® Discover.