Targeted Protein Degradation Services
We offer a variety of services to support targeted protein degradation research, including cell line generation, assay development, screening and profiling services. Learn more about our services below or contact us for details.
Degradation Compound Profiling Services Using Endogenous HiBiT Targets

Endpoint Degradation Profiling
Ideal for screening large compound libraries
- Single concentration or dose response to determine DC50
- Multiple time points to assess fast (5–6 hours) and sustained (18–24 hours) degradation
- Cell viability multiplex


Kinetic Degradation Profiling
Ideal for ranking kinetic parameters of hit degraders
- Single concentration or dose response monitored for 24 hours
- Analysis of degradation rate, Dmax and Dmax50 against concentration to evaluate kinetic potency
- Endpoint cell viability multiplex


Endogenously Tagged HiBiT Neosubstrate Panel to Enable Molecular Glue Discovery

Six neosubstrate targets for IMiD molecular glues were established as HiBiT CRISPR knock-in cell lines across different cell types: IKZF1 and IKZF2 (Jurkat), IKZF3 (MM.1S), SALL 4 (SK-N-DZ), CK1α and GSPT1 (HEK293). This neosubstrate panel enables screening for molecular glue library degradation selectivity and off-target effects with CRBN-recruiting PROTACs. Recommended endpoints for screening include 6 and 24 hours, with concurrent cell viability counter-screen. CC-885 displays broad selectivity, while DKY709 shows robust degradation of IKZF2 and SALL4.
Services for Mechanistic Evaluation and Optimization of Degrader Efficacy
NanoBRET® Target Engagement and Cellular Permeability
- De novo assay and tracer development
- Assessment of degrader binding affinity to target and E3
- Assessment of degrader permeability in live vs permeabilized cells


Degrader Results with Automated VHL TE Assay
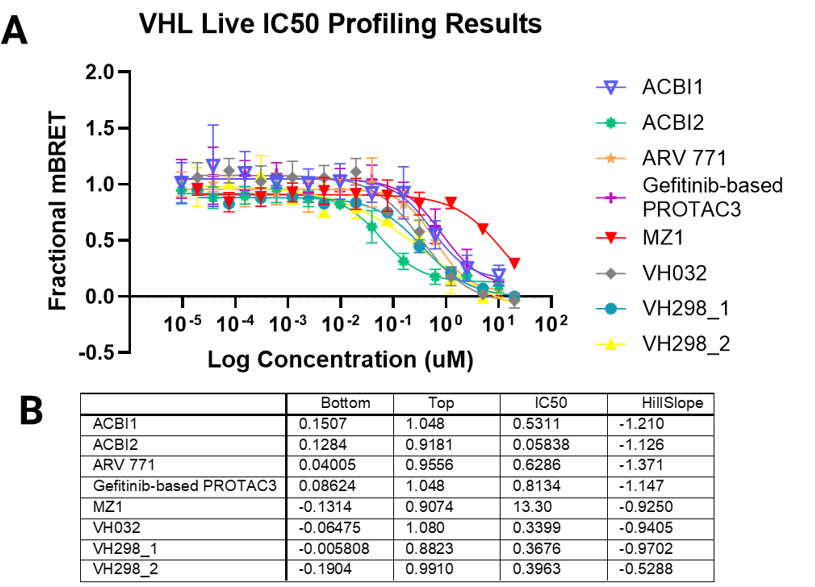
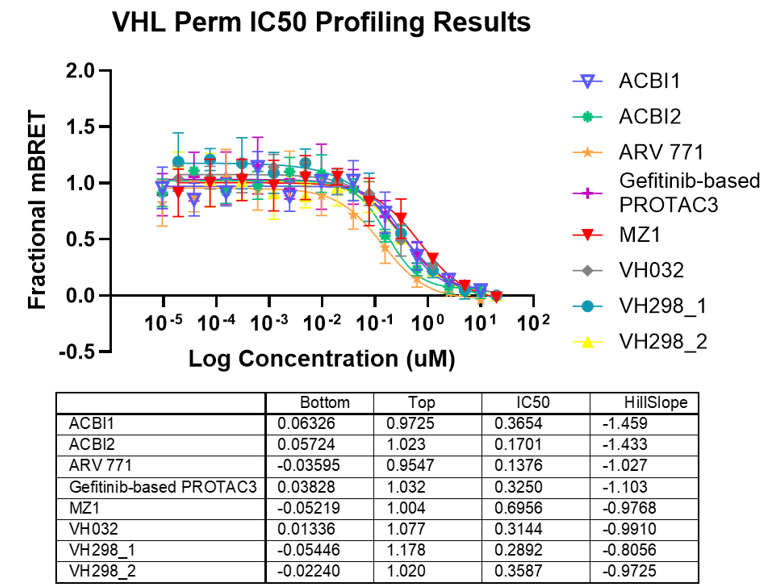
Panel A. Compound concentration-response curves from NanoBRET® TE VHL assays in live-cell or permeabilized-cell modes, respectively. VH298 was used as a permeable control compound. Panel B. Lower and upper asymptote, IC50, and Hill slope parameters calculated from non-linear regression curves.
VHL TE Assay Permeability Parameters
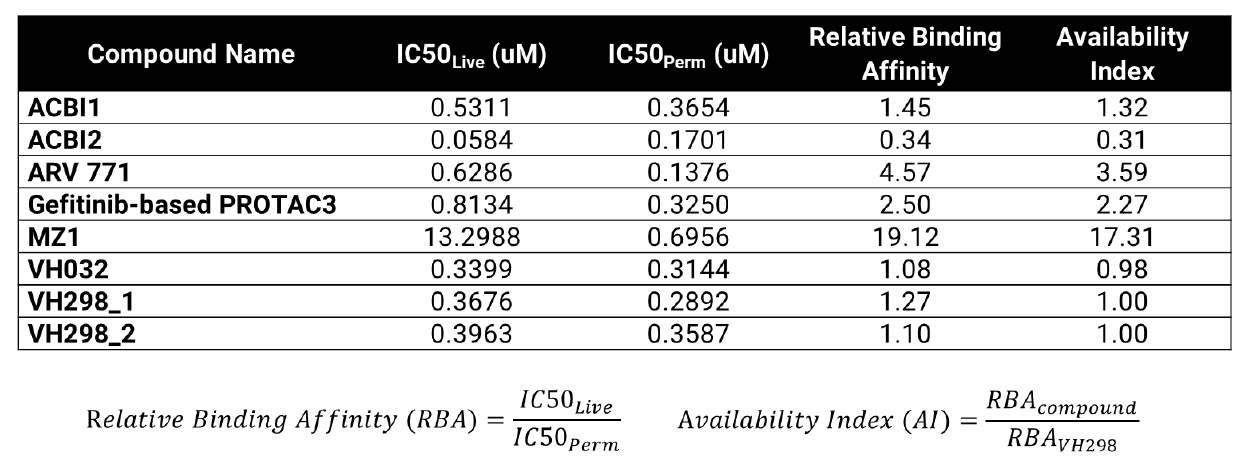
Calculated relative binding affinity (RBA) and availability index (AI) values for each compound. An RBA value of approximately 1 indicates similar free concentrations inside and outside of the cell. AI > 1 suggests reduced availability inside the cell compared to VH298 control. AI < 1 could indicate compound accumulates in cells at higher relative concentrations compared to VH298 control.
NanoBRET® Ternary Complex Formation
- De novo assay development and testing
- Degrader profiling in endpoint or kinetic format
- Single concentration or dose response

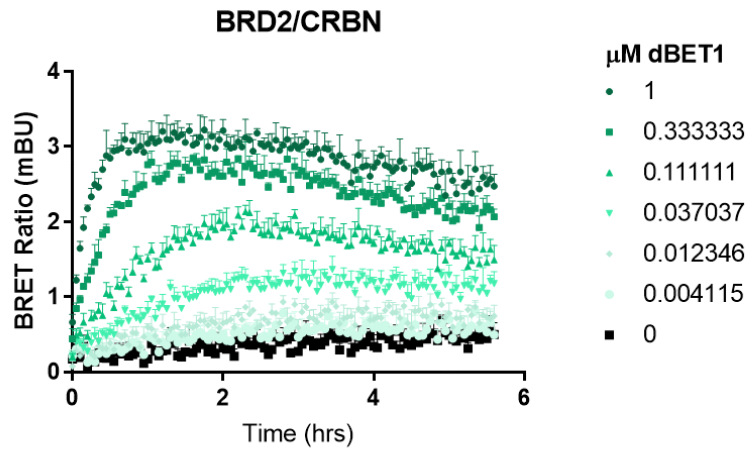
Screening for Neosubstrate Ternary Complex Formation with Transient NanoBRET® Assay


NanoBRET® Ternary Complex Assay optimization for common neosubstrates and CRBN. NanoLuc® neosubstrate and HaloTag® CRBN fusions were expressed in HEK293 cells with or without protease inhibitor MG132. All N- and C- orientations were tested at 1:10 and 1:100 ratios. Results show a significant increase in BRET upon addition of molecular glue due to formation of ternary complex.
NanoBRET® Target Ubiquitination
- De novo assay development and testing
- Degrader profiling in endpoint or kinetic format
- Single concentration or dose response

